Article
Molecular determinants of substrate specificity in human insulin-degrading enzyme
Biochemistry
(2018)
Abstract
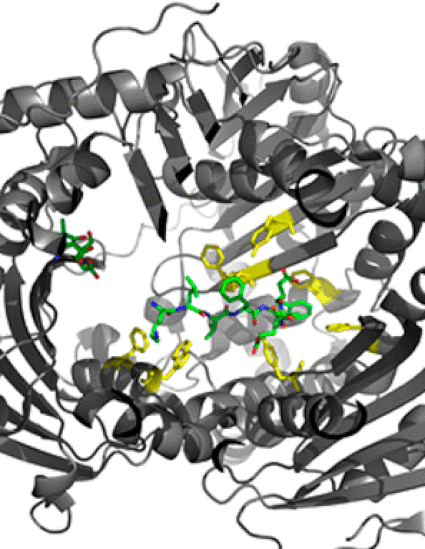
Insulin-degrading enzyme (IDE) is a 110 kDa chambered zinc metalloendopeptidase that degrades insulin, amyloid β, and other intermediate-sized aggregation prone peptides that adopt β-structures. Structural studies of IDE in complex with multiple physiological substrates have suggested a role for hydrophobic and aromatic residues of the IDE active site in substrate binding and catalysis. Here, we examine functional requirements for conserved hydrophobic and aromatic IDE active site residues that are positioned within 4.5 Å of IDE-bound insulin B chain and amyloid β peptides in the reported crystal structures for the respective enzyme-substrate complexes. Charge, size, hydrophobicity, aromaticity, and other functional group requirements for substrate binding IDE active site residues were examined through mutational analysis of the recombinant human enzyme and enzyme kinetic studies conducted using native and fluorogenic derivatives of human insulin and amyloid β peptides. A functional requirement for IDE active site residues F115, A140, F141, Y150, W199, F202, F820, and Y831 was established, and specific contributions of residue charge, size, and hydrophobicity to substrate binding, specificity, and proteolysis were demonstrated. IDE mutant alleles that exhibited enhanced or diminished proteolytic activity toward insulin or amyloid β peptides and derivative substrates were identified.
Disciplines
Publication Date
Summer July 13, 2018
DOI
10.1021/acs.biochem.8b00474
Citation Information
Lazaros Stefanidis, Nicholas D. Fusco, Samantha E. Cooper, Jillian E. Smith-Carpenter, et al.. "Molecular determinants of substrate specificity in human insulin-degrading enzyme" Biochemistry Vol. 57 Iss. 32 (2018) p. 4903 - 4914 Available at: http://works.bepress.com/benjamin_alper/15/
